An introduction to Jupyter - and why I don't like it
2020-11-09
Jupyter Notebooks are hugely popular. In this post I'll give an introduction into what they are, why I would actually not recommend using them, and what I do instead.
I wrote this post for my Python Course
What is Jupyter?
Currently there are two important things:
- Jupyter Notebooks, which are a type of "document" which contains executable code, as well as outputs of that code like images, tables, or text and styled text via Markdown, which also supports mathematical equations.
- Jupyter Lab, which is a browser based IDE.
You can think of a Jupyter notebook like of the ipython console. But instead of typing your code into a terminal, you type it into a code cell in your browser. If you execute it, the results are printed below it and you can then open a new cell to execute further code and still, at any point in time, go back to the above cells and rerun or edit that code easily. If your code creates a plot, that plot will be part of the document too. Here is a screenshot of what this looks like:
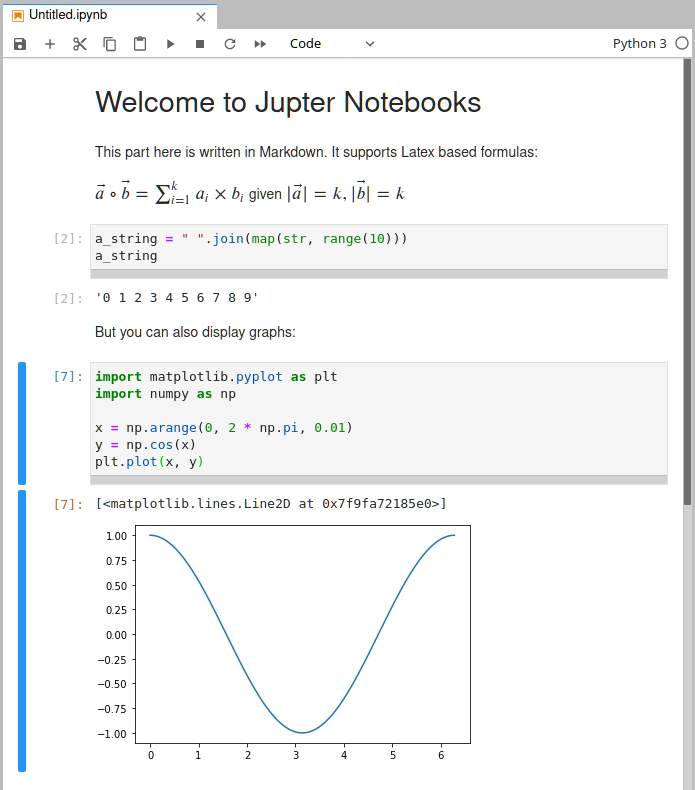
It has many more Features, like interactive sliders, and if you use Jupyter Lab to edit them, it also supports other filetypes, and may look like this:

They are, all in all, pretty damn powerful and extremely popular among data scientists and students. If you do want to use them I'd recommend to follow a tutorial: first Google hit for "jupyter tutorial"
Why I don't like them
I don't want to hate on Jupyter, it's an impressive piece of software that is excellent in what it does; BUT, I just don't think it's a good Idea to do what it does.
Jupyter's main selling point is to combine Code, its results, and text. And there is no point in doing that, except for teaching. Actually for teaching, Jupyter is awesome. However, the most common use case is data analysis. I've used Jupyter Notebooks excessively for nearly two years for the purpose of data analysis and have frequently encountered the following problems:
Crowded Notebooks
Usually, for me, I found that 70% - 90% of the visible area in a notebook was covered with code, rather than with results. To get to a result, you'd have to scroll a lot. Further more, because of the easy access to code that was typed in earlier, I'd regularly go up, make some changes, rerun a cell, and then continue in the bottom. This would lead to notebooks that will not be able to execute from top to bottom because some cells would rely on something that was changed away in a cell above it. Also, I would copy a lot of code between notebooks, which is a terrible thing to do.
Copied Notebooks
Sometimes I did an analysis in Jupyter, and at some point, I would want to run
that same analysis with a different dataset. What would I do? Obviously I'd
copy that notebook, exchange the path in the top, and run it again. (At this
point I'd probably realize, that it can't run from top to bottom without
crashing, so I'd have to spent quite some time on fixing the notebook).
This might happen again. And again. AND THEN you find a bug in that notebook,
and have to fix it in ALL of those pesky copies, <sarcasm>oh the pleasure …
</sarcasm>
Git incompatibility
People tend to have notebooks stored in git repositories, or at least I used to. Sometimes git would see a notebook as changed when I just opened it, and scrolled down without actually making changes. Also notebooks contain images, these Will blow up your repository hugely. Also it's a very bad practice to store data in git. Git is for source code.
You have to edit Stuff in the Browser
Jupyter Lab runs in the browser, this means you have to edit your code in the Browser. I don't like that. I have put a lot of effort into my .vimrc, and I want to use it. I absolutely hate editing Code in notebooks. Yeah, sure, there are plugins that give you some Vim functionality, but that's just additional effort for a worse solution (compared to editing code with Vim)
Conclusion
All of these Points can be countered. You can avoid crowded notebooks by having some self discipline, putting most of your code into python modules that you just call into from the notebook, and it's in your hand whether you check that your notebook stays executable as a whole or not. If you do this, chances are that bugs are in a Python module, and you don't need to edit copied notebooks. You cannot get rid of all the problems that Jupyter notebooks produce within a git repository, but you can solve some of them by deleting all of the outputs before you commit. But if you do this, and also only put very little code into them, because most of your code is in modules, then what is left of the notebooks? Also, this misses another points. Code that generate results, and the results are not the same thing, and don't belong together (at least in my oppinion). Having a script that generates results based on some inputs, and then rerun it, if you want to have an old analysis on new data is a much better practice. Most people that care for your results won't care for your code at all. This way you can also simply send them the results, instead of somehow separating results and code beforehand, but most importantly: you can keep your results out of your git repository. For me the biggest selling point of Jupyter was always how easy it was to get persistent results. Before that I'd manually write code to save a plot as image or a pandas table as csv (ok, that's just one line, but I'm lazy and it annoys me. Additionally, I didn't have one file, where I can also put text, but just mutliple files). And that's the main point: laziness. In the long run, we will tend to take the easiest, most comfortable route, and while it is possible to mitigate a lot of the problems I addressed, it is not the easy thing to do, which means that most of us will, again and again, end up with multiple versions of the same cluttered notebook in a git repository that has a size of 5GB, while 20MB would easily suffice.
I wrote my datasheet library that allows me to store output data of scripts, which tries to emulate the look of Jupyter notebooks without code, easily. For interactive prototyping I use simple plain old python files together with vimteractive (and every other programming editor has a similar functionality). This basically fulfills all the needs of cell based editing that I have. No one expects these scripts to run as a whole afterwards, and I can still keep them around in order to extract some of the prototyped code into tested modules later.
